Please wait while we process your payment
If you don't see it, please check your spam folder. Sometimes it can end up there.
If you don't see it, please check your spam folder. Sometimes it can end up there.
Please wait while we process your payment

By signing up you agree to our terms and privacy policy.
Don’t have an account? Subscribe now
Create Your Account
Sign up for your FREE 7-day trial
By signing up you agree to our terms and privacy policy.
Already have an account? Log in
Your Email
Choose Your Plan
Individual
Group Discount
Save over 50% with a SparkNotes PLUS Annual Plan!
 payment page
payment page
Purchasing SparkNotes PLUS for a group?
Get Annual Plans at a discount when you buy 2 or more!
Price
$24.99 $18.74 /subscription + tax
Subtotal $37.48 + tax
Save 25% on 2-49 accounts
Save 30% on 50-99 accounts
Want 100 or more? Contact us for a customized plan.
 payment page
payment page
Your Plan
Payment Details
Payment Summary
SparkNotes Plus
You'll be billed after your free trial ends.
7-Day Free Trial
Not Applicable
Renews July 25, 2025 July 18, 2025
Discounts (applied to next billing)
DUE NOW
US $0.00
SNPLUSROCKS20 | 20% Discount
This is not a valid promo code.
Discount Code (one code per order)
SparkNotes PLUS Annual Plan - Group Discount
Qty: 00
SparkNotes Plus subscription is $4.99/month or $24.99/year as selected above. The free trial period is the first 7 days of your subscription. TO CANCEL YOUR SUBSCRIPTION AND AVOID BEING CHARGED, YOU MUST CANCEL BEFORE THE END OF THE FREE TRIAL PERIOD. You may cancel your subscription on your Subscription and Billing page or contact Customer Support at custserv@bn.com. Your subscription will continue automatically once the free trial period is over. Free trial is available to new customers only.
Choose Your Plan
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.
For the next 7 days, you'll have access to awesome PLUS stuff like AP English test prep, No Fear Shakespeare translations and audio, a note-taking tool, personalized dashboard, & much more!
You’ve successfully purchased a group discount. Your group members can use the joining link below to redeem their group membership. You'll also receive an email with the link.
Members will be prompted to log in or create an account to redeem their group membership.
Thanks for creating a SparkNotes account! Continue to start your free trial.
We're sorry, we could not create your account. SparkNotes PLUS is not available in your country. See what countries we’re in.
There was an error creating your account. Please check your payment details and try again.
Please wait while we process your payment

Your PLUS subscription has expired
Please wait while we process your payment
Please wait while we process your payment

Prokaryotic Initiation
In prokaryotic cells, free RNA polymerase molecules are constantly colliding with DNA helices. The collision leads to a weak association between the DNA and RNA polymerase, which is soon broken. However, when the RNA polymerase binds to a specific sequence on the DNA, it binds tightly, forming a DNA/RNA polymerase complex. This specific site of binding is called the start site. The start site represents the location on the DNA that marks where the first nucleotide of an RNA chain should go; that spot is designated as the "plus one position". Positions that are designated as downstream in the RNA are positively numbered according to their relative position to the plus one position. All positions designated as upstream of the start site are labeled with negative numbers according to their position relative to the start site. Sequences located just upstream of the start site, called the promoter region, contain the information that signals the RNA polymerase to start transcription.
There are a number of key features to the promoter region that give it the ability to provide the signal initiating transcription. While nearly all promoters vary slightly, they all have general traits that can be identified. Located approximately 10 and 32 base pairs upstream of the start site are two such regions, called the -10 and -35 sequences. Each sequence consists of six base pairs. For an ideal promoter, the sequence is TTGACA for the -35 region and TATAAT for the -10 region.
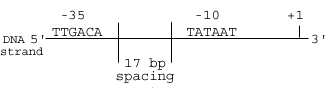
In addition to the specificity of the bases in these sequences, the spacing between the two is also important. Ideally, this gap is 17 base pairs long. Deviations from this spacing have significant effects on the strength of the promoter region. The closer a promoter region is to matching this canonical promoter sequence, the greater its strength.
There is a third promoter element that is sometimes seen in very strong promoters which is called the UP element. It usually is composed of alternating stretches of 5 adenine and thymine bases. It is located upstream of the -35 region.
RNA polymerase binds to the DNA helix at the start site. Bound to DNA, it covers a 60 base pair region within which it scans for the -35 and -10 promoters. Initially, the polymerase, and specifically the sigma subunit, binds in what is called a "closed complex" to the DNA. The RNA polymerase/promoter complex then undergoes a conformational change that breaks a number of base pairs extending from the -10 region to create a bubble in which the two DNA strands have separated. This bubble is usually approximately 17 base pairs in length. This new formation is called the "open complex". RNA synthesis is then initiated using one of the DNA strands as a template for adding complementary RNA base pairs. Transcription is usually initiated with a purine, rather than pyrimidine, base. Once initiated, the RNA polymerase moves down the DNA strand in the elongation process, which is covered in the next section.
Please wait while we process your payment

