Please wait while we process your payment
If you don't see it, please check your spam folder. Sometimes it can end up there.
If you don't see it, please check your spam folder. Sometimes it can end up there.
Please wait while we process your payment

By signing up you agree to our terms and privacy policy.
Don’t have an account? Subscribe now
Create Your Account
Sign up for your FREE 7-day trial
By signing up you agree to our terms and privacy policy.
Already have an account? Log in
Your Email
Choose Your Plan
Individual
Group Discount
Save over 50% with a SparkNotes PLUS Annual Plan!
 payment page
payment page
Purchasing SparkNotes PLUS for a group?
Get Annual Plans at a discount when you buy 2 or more!
Price
$24.99 $18.74 /subscription + tax
Subtotal $37.48 + tax
Save 25% on 2-49 accounts
Save 30% on 50-99 accounts
Want 100 or more? Contact us for a customized plan.
 payment page
payment page
Your Plan
Payment Details
Payment Summary
SparkNotes Plus
You'll be billed after your free trial ends.
7-Day Free Trial
Not Applicable
Renews July 7, 2025 June 30, 2025
Discounts (applied to next billing)
DUE NOW
US $0.00
SNPLUSROCKS20 | 20% Discount
This is not a valid promo code.
Discount Code (one code per order)
SparkNotes PLUS Annual Plan - Group Discount
Qty: 00
SparkNotes Plus subscription is $4.99/month or $24.99/year as selected above. The free trial period is the first 7 days of your subscription. TO CANCEL YOUR SUBSCRIPTION AND AVOID BEING CHARGED, YOU MUST CANCEL BEFORE THE END OF THE FREE TRIAL PERIOD. You may cancel your subscription on your Subscription and Billing page or contact Customer Support at custserv@bn.com. Your subscription will continue automatically once the free trial period is over. Free trial is available to new customers only.
Choose Your Plan
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.
For the next 7 days, you'll have access to awesome PLUS stuff like AP English test prep, No Fear Shakespeare translations and audio, a note-taking tool, personalized dashboard, & much more!
You’ve successfully purchased a group discount. Your group members can use the joining link below to redeem their group membership. You'll also receive an email with the link.
Members will be prompted to log in or create an account to redeem their group membership.
Thanks for creating a SparkNotes account! Continue to start your free trial.
We're sorry, we could not create your account. SparkNotes PLUS is not available in your country. See what countries we’re in.
There was an error creating your account. Please check your payment details and try again.
Please wait while we process your payment

Your PLUS subscription has expired
Please wait while we process your payment
Please wait while we process your payment

Overview of Post-Transcriptional RNA Splicing
Prokaryotic DNA transcription produces messenger RNA, which is necessary for transfer from the cell nucleus to the cytoplasm where translation occurs. In contrast, eukaryotic DNA transcription takes place in a cell's nucleus and produces what is called a primary RNA transcript or pre-messenger RNA. Before eukaryotic products of transcription can be moved into the cytoplasm, they must undergo modifications that allow them to become mature messenger RNA. Splicing is the name given to the reaction that removes unnecessary segments of the primary RNA transcript, called introns. The removal of the introns produces mRNA (see the figure, below). Messenger RNA contains only exons, those portions of the primary RNA transcript that will be translated into a protein.
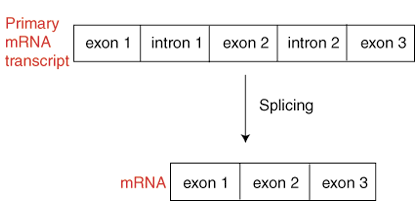
Unlike the sequence of an exon, intron sequences are unimportant. Only small portions of an intron sequence are preserved. These portions, located near the end of each intron, serve to identify a sequence as an intron, identifying the sequence for removal. There are intron identifying portions:
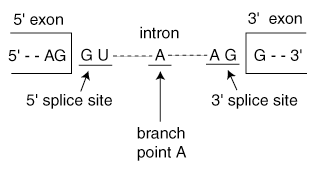
With the help of the spliceosome, a multi-component protein, the splicing reaction occurs in two steps. The spliceosome contains five small nuclear ribonucleoproteins (snRNPS, pronounced "snurps"). They are called U1, U2, U4, U5, and U6. Each snRNP contains protein components that are critical for the splicing reaction. U1 binds directly to the 5' splice site via complementary base pairing. U1 then recruits U2, which forms a complex with branch point A. U4 and U6 work in concert to form a "pre-splicing complex" and U5 helps to hold the exons in place between the first and second steps in the splicing reaction. Once the splicing reactions have occurred and the exons have been joined, the resulting mRNA is freed from the spliceosome machinery and the different snRNP components are recycled for further use.
In addition to the post-transcriptional modifications already discussed (5' cap, poly A tail addition, and splicing), a fourth type of modification can be made: RNA editing. RNA editing is a modification that changes the mRNA sequence and as a result alters the protein produced by that mRNA. Editing can occur in two ways. First, by changing one nucleotide to another, and second by inserting or deleting a nucleotide or nucleotides.
Please wait while we process your payment

