Please wait while we process your payment
If you don't see it, please check your spam folder. Sometimes it can end up there.
If you don't see it, please check your spam folder. Sometimes it can end up there.
Please wait while we process your payment

By signing up you agree to our terms and privacy policy.
Don’t have an account? Subscribe now
Create Your Account
Sign up for your FREE 7-day trial
By signing up you agree to our terms and privacy policy.
Already have an account? Log in
Your Email
Choose Your Plan
Individual
Group Discount
Save over 50% with a SparkNotes PLUS Annual Plan!
 payment page
payment page
Purchasing SparkNotes PLUS for a group?
Get Annual Plans at a discount when you buy 2 or more!
Price
$24.99 $18.74 /subscription + tax
Subtotal $37.48 + tax
Save 25% on 2-49 accounts
Save 30% on 50-99 accounts
Want 100 or more? Contact us for a customized plan.
 payment page
payment page
Your Plan
Payment Details
Payment Summary
SparkNotes Plus
You'll be billed after your free trial ends.
7-Day Free Trial
Not Applicable
Renews July 19, 2025 July 12, 2025
Discounts (applied to next billing)
DUE NOW
US $0.00
SNPLUSROCKS20 | 20% Discount
This is not a valid promo code.
Discount Code (one code per order)
SparkNotes PLUS Annual Plan - Group Discount
Qty: 00
SparkNotes Plus subscription is $4.99/month or $24.99/year as selected above. The free trial period is the first 7 days of your subscription. TO CANCEL YOUR SUBSCRIPTION AND AVOID BEING CHARGED, YOU MUST CANCEL BEFORE THE END OF THE FREE TRIAL PERIOD. You may cancel your subscription on your Subscription and Billing page or contact Customer Support at custserv@bn.com. Your subscription will continue automatically once the free trial period is over. Free trial is available to new customers only.
Choose Your Plan
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.
For the next 7 days, you'll have access to awesome PLUS stuff like AP English test prep, No Fear Shakespeare translations and audio, a note-taking tool, personalized dashboard, & much more!
You’ve successfully purchased a group discount. Your group members can use the joining link below to redeem their group membership. You'll also receive an email with the link.
Members will be prompted to log in or create an account to redeem their group membership.
Thanks for creating a SparkNotes account! Continue to start your free trial.
We're sorry, we could not create your account. SparkNotes PLUS is not available in your country. See what countries we’re in.
There was an error creating your account. Please check your payment details and try again.
Please wait while we process your payment

Your PLUS subscription has expired
Please wait while we process your payment
Please wait while we process your payment

DNA Proof-Reading and Repair
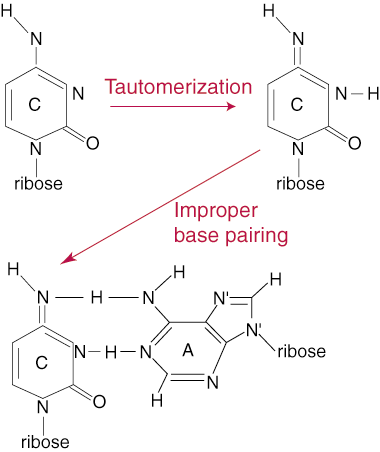
The low overall rate of mutation during DNA replication (1 base pair change in one billion base pairs per replication cycle) does not reflect the true number of errors that take place during the replication process. The number is kept so low by a proof-reading system that checks newly synthesized DNA for errors and corrects them when they are found. Errors in DNA replication can take different forms, but usually revolve around the addition of a nucleotide with the incorrect base, meaning the pairing between the parent and daughter strand bases is not complementary. The addition of an incorrect base can take place by a process called tautomerization. A tautomer of a base group is a slight rearrangement of its electrons that allows for different bonding patterns between bases. This can lead to the incorrect pairing of C with A instead of G, for example.
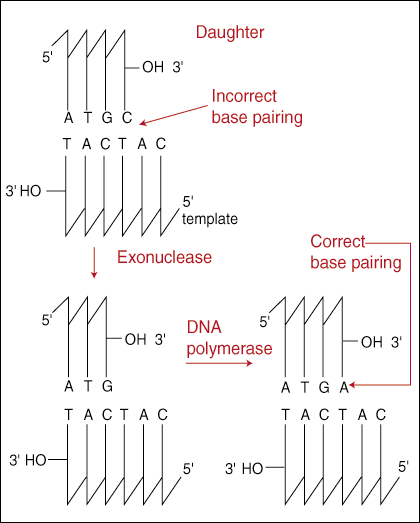
DNA retains its high level of accuracy is with its proof-reading function.
The 3' to 5' proof-reading exonuclease works by scanning along directly behind as the DNA polymerase adds new nucleotides to the growing strand. If the last nucleotide added is mismatched, then the entire replication holoenzyme backs up, removes the last incorrect base, and attempts to add the correct base again. The enzyme is "3' to 5'" because it scans in the opposite direction of DNA replication, which we learned must always be 5' to 3'. The mechanism of the proof-reading system offers an explanation as to why DNA replication must occur in this direction.
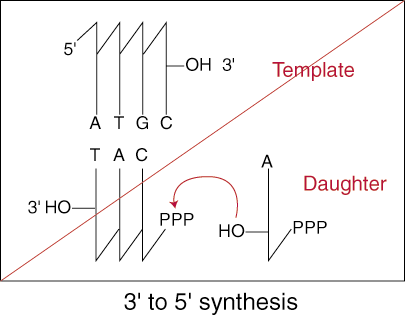
Keeping in mind the chemical mechanism we learned for the addition of
nucleotides to the growing DNA strand, imagine what happens when the proof-
reading system removes an incorrectly paired base. The exonuclease removes the
base by cleaving the phosphodiester bond that had just been formed. In 5' to 3'
synthesis, this leaves the 3' -OH still attached to the terminal end of the
growing strand ready to attack another nucleotide.
After DNA has been completely replicated, the daughter strand is often not a perfect copy of the parent strand it came from. Mutations during replication and damage after replication make it necessary for there to be a repair system to fix any errors in newly synthesized DNA. There are three main sources of damage to DNA.
These different sources of damage lead to different categories of DNA damage. The damage that is caused by water attack can lead to unnatural bases. Chemical and radiation damage leads to the formation of bulky adducts to, or breaks in, the growing DNA strand. In the previous section we discussed the 3' to 5' proof-reading exonuclease that is responsible for fixing mismatches. Because it is not a perfect system, it can miss mismatched bases. As a result, a third category of DNA damage is mismatched bases.
Please wait while we process your payment

